Abstract Differential expression analysis in single-cell transcriptomics enables the dissection of cell-type-specific responses to perturbations such as disease, trauma, or experimental manipulations. While many statistical methods are available to identify differentially expressed genes, the principles that distinguish these methods and their performance remain unclear. Here, we show that the relative performance of these methods is contingent on their ability to account for variation between biological replicates. Methods that ignore this inevitable variation are biased and prone to false discoveries. Indeed, the most widely used methods can discover hundreds of differentially expressed genes in the absence of biological differences. To exemplify these principles, we exposed true and false discoveries of differentially expressed genes in the injured mouse spinal cord.
Publication scientifique
Confronting false discoveries in single-cell differential expression
Autres publications de la plateforme
Hypothalamic deep brain stimulation augments walking after spinal cord injury
A neuronal architecture underlying autonomic dysreflexia
Dual lineage origins contribute to neocortical astrocyte diversity
Urolithin A provides cardioprotection and mitochondrial quality enhancement preclinically and improves...
Regional differences in progenitor metabolism shape brain growth during development
Journal de publication
Auteurs:
Date de publication:
Plateforme:
Études récentes de la plateforme

Restaurer le mouvement après une paralysie
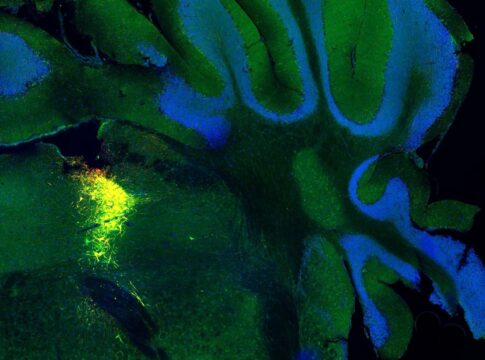
Décoder les ondes de l’activité cérébrale